
April 14, 2016
Salk team’s improved rabies virus technology maps neurons across large swaths of nervous system
Salk team’s improved rabies virus technology maps neurons across large swaths of nervous system
LA JOLLA—Salk Institute scientists have developed a new reagent to map the brain’s complex network of connections that is 20 times more efficient than their previous version. This tool improves upon a technique called rabies virus tracing, which was originally developed in the Callaway lab at Salk and is commonly used to map neural connections.
Rabies viral tracing uses a modified version of the rabies virus that jumps between neurons, lighting up connections along the way. The illuminated map allows researchers to precisely trace which neurons connect to each other. Visualizing this neural circuitry can help scientists learn more about conditions ranging from motor diseases to neurodevelopmental disorders.
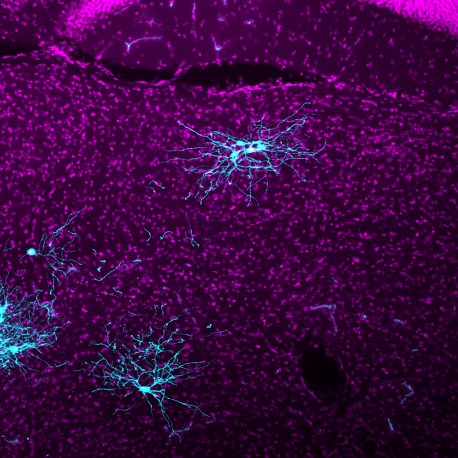
Click here for a high-resolution image.
Credit: Salk Institute
“To truly understand brain function, we have to understand how different types of neurons are connected to each other across many distant brain areas. The rabies tracing methods we have developed made that possible, but we were only labeling a fraction of all of the connections,” says Edward Callaway, a Salk professor and senior author of the new paper, published April 14, 2016 in the journal Cell Reports.
He adds that such a dramatic improvement in a critical tool for neuroscience will help researchers illuminate aspects of brain disorders where connectivity and global processing goes awry, such as in autism and schizophrenia.
Long distance connections between neurons are key to what is called global processing in the brain. Imagine a ball sailing toward a catcher. The catcher’s visual circuits will process the information about the ball and send that information over to the brain’s motor circuits. The motor circuits then direct nerves in the catcher’s arm and hand to grab the ball. That global processing relies on long-distance neural circuits forming precise connections to specific neuron types; these circuits can be revealed with rabies viral tracers.
“With this new rabies tracer, we can visualize connectivity neuron by neuron, and across long distance input neurons better than with previous rabies tracers,” says Euiseok Kim, a Salk research associate and first author of the paper.
There are billions of neurons in the brain, and only a handful of technologies that can map the communication going on between them. Some imaging techniques such as functional MRIs can visualize broad scale communication across the brain, but do not focus on the cellular level. Electrophysiology and electron microscopy can track cell-to-cell connectivity, but aren’t suited to mapping neural circuits across the whole brain.
Tracing methods using neurotropic viruses, like rabies, have long been utilized to trace connections across neural pathways. But these viruses spread widely throughout the brain across multiple circuits, making it difficult to determine which neurons are directly connected. In 2007, Callaway’s lab pioneered a new approach based on genetically modified rabies virus. This approach allowed the viral infection to be targeted to specific types of neurons and also allowed the spread of the virus to be controlled. The result is that this system illuminates neurons across the entire brain, but labels only those that are directly connected to neurons of interest.
Click here
for a high-resolution imageCredit: Salk Institute
To control how far the virus travels, scientists ensure the rabies virus can only infect a select group of neurons. First scientists remove and replace the crucial outer-coat of the rabies virus, called glycoproteins. The virus needs this coat of glycoproteins to enter and infect cells, but the replacement glycoprotein prevents the virus from infecting normal neurons. Scientists then alter a group of neurons in mice to become so-called “starter cells” that are uniquely susceptible to infection with the modified glycoprotein. Starter cells are also programmed to provide the rabies glycoproteins so that once a starter cell is infected, new copies of the rabies tracer can spread across the starter cell’s synapses into connected neurons. However, once the rabies viral tracer is in the next set of neurons, it won’t find the glycoprotein it needs to continue to spread, and so the trail of infection across neural circuits ends.
Although the original rabies viral tracer accurately traces circuits, it was only crossing a fraction of the starter cell’s synapses. The Salk research team went about engineering a more efficient rabies viral tracer. First, the researchers took pieces from various rabies strains to create new chimeric glycoproteins and then tested the versions in by counting labeled cells in known circuits.
The winning chimeric glycoprotein was further genetically modified with a technique called codon optimization to increase levels of the glycoprotein produced in starter cells. Compared to the original rabies tracer, the new codon-optimized tracer increased the tracing efficiency for long distance input neurons by up to 20 fold.
“Although this improved version is much better, there are still opportunities to improve the rabies tracer further as we continue to examine other rabies strains,” says Kim.
Other co-authors of the study were Matthew Jacobs and Tony Ito-Cole of the Salk Institute’s Systems Neurobiology Laboratory.
This work was funded in part by the National Institutes of Health, Gatsby Charitable Foundation and the Howard Hughes Medical Institute Collaborative Innovation Award.
JOURNAL
Cell Reports
AUTHORS
Euiseok J. Kim, Matthew W. Jacobs, Tony Ito-Cole, Edward M. Callaway
Office of Communications
Tel: (858) 453-4100
press@salk.edu
Unlocking the secrets of life itself is the driving force behind the Salk Institute. Our team of world-class, award-winning scientists pushes the boundaries of knowledge in areas such as neuroscience, cancer research, aging, immunobiology, plant biology, computational biology and more. Founded by Jonas Salk, developer of the first safe and effective polio vaccine, the Institute is an independent, nonprofit research organization and architectural landmark: small by choice, intimate by nature, and fearless in the face of any challenge.