
April 7, 2015
Salk scientists discover a single protein that energizes both muscles and the brain
Salk scientists discover a single protein that energizes both muscles and the brain
LA JOLLA–Just as some people seem built to run marathons and have an easier time going for miles without tiring, others are born with a knack for memorizing things, from times tables to trivia facts. These two skills–running and memorizing–are not so different as it turns out.
Salk scientists and collaborators have discovered that physical and mental activities rely on a single metabolic protein that controls the flow of blood and nutrients throughout the body, as reported in the journal Cell Metabolism. The new study could point to potential treatments in regenerative and developmental medicine as well as ways to address defects in learning and memory.
“This is all about getting energy where it’s needed to ‘the power plants’ in the body,” says Ronald Evans, director of Salk’s Gene Expression Laboratory and senior author of the new paper, published April 7, 2015. “The heart and muscles need a surge of energy to carry out exercise and neurons need a surge of energy to form new memories.”
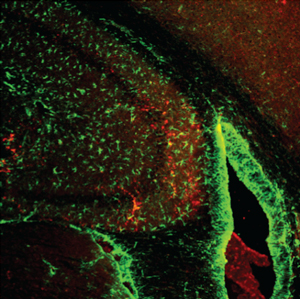
Salk researchers and collaborators discovered that physical and mental activities rely on a single metabolic protein, ERRγ, that controls the flow of blood and nutrients throughout the body. In this image, ERRγ is shown (stained red) in the hippocampus, the area of the brain largely responsible for memory. The new work could point to a way to enhance learning.
Click here for a high-resolution image.
Image: Courtesy of the Salk Institute for Biological Studies
Energy for muscles and brains, the scientists discovered, is controlled by a single protein called estrogen-related receptor gamma (ERRγ). Evans’ research group has previously studied the role of ERRγ in the heart and skeletal muscles. In 2011, they discovered that promoting ERRγ activity in the muscle of sedentary mice increased blood supply to their muscles and doubled their running capacity. ERRγ, they went on to show, turns on a whole host of muscle genes that convert fat to energy.
Thus, ERRγ became known as a master metabolic switch that energized muscle to enhance performance. Although studies had also shown that ERRγ was active in the brain, researchers didn’t understand why–the brain burns sugar and ERRγ was previously shown to only burn fat. So the team decided to look more closely at what the protein was doing in brain cells.
By first looking at isolated neurons, Liming Pei, lead and co-corresponding author of the paper, found that, as in muscle, ERRγ activates dozens of metabolic genes in brain cells. Unexpectedly, this activation related to sugar instead of fat. Neurons that lacked ERRγ could not ramp up energy production and thus had a compromised performance.
“We assumed that ERRγ did the same thing throughout the body,” says Evans. “But we learned that it’s different in the brain.” ERRγ, they now conclude, turns on fat-burning pathways in muscles and sugar-burning pathways in the brain.
Evans and his collaborators noticed that ERRγ in live mice was most active in the hippocampus–an area of the brain that is active in producing new brain cells, is involved in learning and memory and is known to require lots of energy. They wondered whether ERRγ had a direct role in learning and memory. By studying mice lacking ERRγ in the brain, they found a link.
While mice without the protein had normal vision, movement and balance, they were slower at learning how to swim through a water maze–and poor at remembering the maze on subsequent trials–compared to mice with normal levels of ERRγ.
“What we found is that mice that missing ERRγ are basically very slow learners,” says Pei. Varying levels of ERRγ could also be at the root of differences between how individual humans learn, he hypothesizes. “Everyone can learn, but some people learn and memorize more efficiently than others, and we now think this could be linked to changes in brain metabolism.”
A better understanding of the metabolism of neurons could help point the way to improved treatments for learning and attention disorders. And possibly, revving up levels of ERRγ could even enhance learning, just as it enhances muscle function.
“What we’ve shown is that memories are really built on a metabolic scaffold,” says Evans. “And we think that if you want to understand learning and memory, you need to understand the circuits that underlie and power this process.”
Other researchers on the study were Yangling Mu, Mathias Leblanc, William Alaynick, Matthew Pankratz, Tiffany W. Tseng, Samantha Kaufman, Ruth T. Yu, Michael Downes, Samuel L. Pfaff, and Fred H. Gage, all of the Salk Institute for Biological Studies; Liming Pei of the University of Pennsylvania; Grant D. Barish of Northwestern University; Christopher Liddle of the University of Sydney; and Johan Auwerx of Ecole Polytechnique Federale de Lausanne.
The work was supported by the Howard Hughes Medical Institute, the National Institutes of Health, the Leona M. and Harry B. Helmsley Charitable Trust, the Ellison Medical Foundation and Glenn Foundation for Medical Research, the Children’s Hospital of Philadelphia and the
Penn Medicine Neuroscience Center.
About the Salk Institute for Biological Studies:
The Salk Institute for Biological Studies is one of the world’s preeminent basic research institutions, where internationally renowned faculty probes fundamental life science questions in a unique, collaborative and creative environment. Focused both on discovery and on mentoring future generations of researchers, Salk scientists make groundbreaking contributions to our understanding of cancer, aging, Alzheimer’s, diabetes and infectious diseases by studying neuroscience, genetics, cell and plant biology, and related disciplines.
Faculty achievements have been recognized with numerous honors, including Nobel Prizes and memberships in the National Academy of Sciences. Founded in 1960 by polio vaccine pioneer Jonas Salk, MD., the Institute is an independent nonprofit organization and architectural landmark.
JOURNAL
Cell Metabolism
AUTHORS
Yangling Mu, Mathias Leblanc, William Alaynick, Matthew Pankratz, Tiffany W. Tseng, Samantha Kaufman, Ruth T. Yu, Michael Downes, Samuel L. Pfaff, Fred H. Gage, and Ronald M. Evans of the Salk Institute for Biological Studies; Liming Pei of the University of Pennsylvania; Grant D. Barish of Northwestern University; Christopher Liddle of the University of Sydney; and Johan Auwerx of Ecole Polytechnique Federale de Lausanne
Office of Communications
Tel: (858) 453-4100
press@salk.edu