
July 1, 2020
Stool microbes make a challenging diagnosis easier
Stool microbes make a challenging diagnosis easier
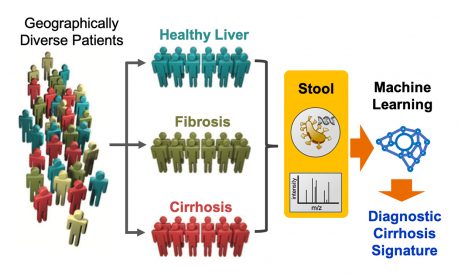
LA JOLLA—Chronic liver disease represents a major global public health problem affecting an estimated 844 million people, according to the World Health Organization. It is among the top causes of mortality in Australia, the UK and the United States. At the same time, it is both difficult to manage and there is no FDA-approved anti-fibrotic liver therapy. The microbiome—a complex collection of microbes that inhabit the gut—may be an unexpected indictor of health. Now, a collaborative team of Salk Institute and UC San Diego scientists have created a novel microbiome-based diagnostic tool that, with the accuracy of the best physicians, quickly and inexpensively identifies liver fibrosis and cirrhosis over 90 percent of the time in human patients.
Click here for a high-resolution image.
Credit: Salk Institute
The non-invasive method relies on an algorithm to analyze patient stool samples—which contains traces of what lives in the gut—and could lead to improved patient care and treatment outcomes for liver disease, as detailed online on June 30, 2020 in Cell Metabolism.
“The microbiome is a dynamic living sensor of small changes in health and disease in the body, and as such, it provides an accurate readout of body health,” says Salk Professor Ronald Evans, co-corresponding author and holder of the March of Dimes Chair. “Because this diagnostic is fast and low-cost, it could be something that becomes widely used, especially in the many areas that lack specialty clinics and physicians. Simply said, it could be a real game changer, with world-wide implications.”
Non-alcoholic fatty liver disease (NAFLD) is the leading cause of chronic liver disease globally and can progress to liver fibrosis and cirrhosis and potentially cancer, as the liver starts to experience scarring and cell death. But diagnostic tools for liver fibrosis and cirrhosis are lacking. Biopsies are invasive and can miss injured regions of the liver, and MRIs are expensive and are often not available in rural areas. To address these challenges, the research team explored the microbiome as a way to meet the urgent need for a new test to identify patients at risk.
“We sought to develop a universal, non-invasive test for liver fibrosis and cirrhosis based on a ‘microbiome signature’ of the disease,” says Michael Downes, a Salk senior staff scientist and co-author of the study.
In collaboration with scientists from the UC San Diego Department of Medicine, the team optimized a computational method called machine learning to uncover a complex disease signature based on 19 bacterial species present in the stool samples of a patient group. The signature is made up of the different quantities of bacteria, creating a universal fingerprint for identifying liver fibrosis and cirrhosis. The study included 163 clinical samples from both healthy as well as sick family members to identify variables that were indicative of liver disease.
Using data from microbiome genetic profiling and from metabolites from the stool samples, the researchers discovered a microbiome signature that was associated with a cirrhosis diagnosis with 94 percent accuracy. The microbiome signature could also determine the stage of liver fibrosis, which could allow doctors to grade patients based on their stage of the disease and improve treatment strategies.
“These findings demonstrate that it is possible to use machine learning to identify a universal signature that can be used for accurate diagnosis of a disease, such as liver cirrhosis,” says Tae Gyu Oh, first author of the paper and a postdoctoral researcher in the Evans lab. “The patterns we found reflect the complexity of the microbiome and how gut health likely affects disease.”

Click here for a high-resolution image.
Credit: Salk Institute
The researchers then applied their microbiome signature to two independent populations of patients from China and Italy. The team’s signature could accurately identify cirrhosis in over 90 percent of patients, which validates the power and accuracy of the algorithm across different genetics and diets.
“It is remarkable that a gut microbiome signature derived from patients residing in Southern California for cirrhosis was able to predict cirrhosis in two independent cohorts residing in China and Italy. It speaks to the new discoveries that are yet to be realized in the role of the gut microbiome to diagnose and risk-stratify liver disease,” says Rohit Loomba, co-corresponding author and director of the NAFLD Research Center at the UC San Diego School of Medicine. “I think the power of using the microbiome as a diagnostic tool is only starting to be realized.”
In the future, the scientists will examine the causal link between the microbiome and liver disease by testing whether restoring parts of the microbiome leads to regression of the disease or removing certain bacteria makes it worse. The team also hopes this approach can be used to characterize additional diseases, such as inflammatory bowel disease, colon cancer, Alzheimer’s and other diseases shown to be likely affected by a dysregulated microbiome.
Other researchers on the paper were Ting Fu, Ruth T. Yu and Annette R. Atkins of Salk; Susy M. Kim, Cyrielle Caussy, Shirin Bassirian, Seema Singh, Egbert V. Madamba, Ricki Bettencourt, Lisa Richards, Manuela Raffatellu, Pieter C. Dorrestein, David A. Brenner, Claude B. Sirlin and Rob Knight of UC San Diego; and Jian Guo and Tao Huan of the University of British Columbia Faculty of Science.
This work was supported by grants from the National Institutes of Health (P42ES010337, HL088093, DK057978, HL105278, ES010337); the Howard Hughes Medical Institute; the Salk Institute Cancer Center (CA014195); the NOMIS Foundation; the Fondation Leducq; Public Health Service Grants (AI126277, AI114625, AI145325); the Chiba University-UCSD Center for Mucosal Immunology, Allergy, and Vaccines; the UCSD Department of Pediatrics; the Burroughs Wellcome Fund; NIEHS (5P42ES010337); NCATS (5UL1TR001442); NIDDK (U01DK061734, R01DK106419, P30DK120515, R01DK121378); and DOD PRCRP (W81XWH-18-2-0026). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
DOI: 10.1016/j.cmet.2020.06.005
JOURNAL
Cell Metabolism
AUTHORS
Tae Gyu Oh, Susy M. Kim, Cyrielle Caussy, Ting Fu, Jian Guo, Shirin Bassirian, Seema Singh, Egbert V. Madamba, Ricki Bettencourt, Lisa Richards, Manuela Raffatellu, Pieter C. Dorrestein, Ruth T. Yu, Annette R. Atkins, Tao Huan, David A. Brenner, Claude B. Sirlin, Rob Knight, Michael Downes, Ronald M. Evans and Rohit Loomba
Office of Communications
Tel: (858) 453-4100
press@salk.edu
Unlocking the secrets of life itself is the driving force behind the Salk Institute. Our team of world-class, award-winning scientists pushes the boundaries of knowledge in areas such as neuroscience, cancer research, aging, immunobiology, plant biology, computational biology and more. Founded by Jonas Salk, developer of the first safe and effective polio vaccine, the Institute is an independent, nonprofit research organization and architectural landmark: small by choice, intimate by nature, and fearless in the face of any challenge.
Notifications