
May 15, 2009
Missing genomic "fence posts" explain inactivated tumor suppressor genes in breast cancer
Missing genomic "fence posts" explain inactivated tumor suppressor genes in breast cancer
La Jolla, CA-Our genome is a patchwork of neighborhoods that couldn’t be more different: Some areas are hustling and bustling with gene activity, while others are sparsely populated and in perpetual lock-down. Breaking down just a few of the molecular fences that separate them blurs the lines and leads to the inactivation of at least two tumor suppressor genes, according to researchers at the Salk Institute for Biological Studies.
Their findings, published in the May 15, 2009 issue of Molecular Cell, explain how a single event can put a cell well ahead on the road to becoming a tumor cell. “Selectively removing a couple of fence posts jumpstarts a cascade of global changes all over the genome that may eventually lead to cancer,” says Beverly Emerson, Ph.D., a professor in the Regulatory Biology Laboratory, who led the study.
Normally, a complex network of accelerators (growth factors) and brakes (tumor suppressors) keeps a tight lid on cell proliferation. Tumors result when changes in the genome activate cancer-causing genes or inactivate tumor suppressor genes that tip this delicate balance in favor of uncontrolled cell growth.
“For a really long time people have been trying to understand how tumor suppressor genes get silenced in cancer,” says postdoctoral researcher and first author Michael Witcher. “Now that we have figured out one of the key events that leads to their inactivation, we might be able to exploit this mechanism to develop novel therapies.”
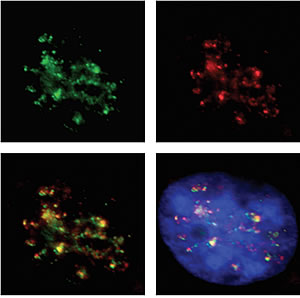
Top left: Molecular fence posts (shown in green) separate active from inactive chromosomal neighborhoods areas. Top right: RNA polymerase 2 (shown in red) reads genes in active neighborhoods. Bottom left: Intact fences co-localize with areas of gene activity. Areas of overlap show up in yellow. Bottom right: DNA is shown in blue.
Image: Courtesy of Michael Witcher, Salk Institute for Biological Studies
If stretched out, the DNA of a single human cell would form a very thin thread about 6 feet in length. To fit such a long molecule inside a cell’s nucleus and keep everything neatly organized, the DNA is threaded around histone proteins and coiled up in a highly condensed structure called heterochromatin. In areas of gene activity, the tightly packed chromatin is unfurled just enough to make the DNA accessible to regulatory proteins.
In many different types of cancers, however, including breast, lung, liver, and pancreatic tumors, as well as multiple myeloma and lymphoma, the tumor suppressor p16 gets buried deep inside heterochromatin. As a result, it cannot be read by the transcription machinery and is unable keep watch over cell growth.
Researchers had known for a long time that sometimes p16 is silenced long before a cell turns cancerous, yet why that particular stretch of DNA was flagged with chemical marks and became wound up so tightly that it became inaccessible had remained a mystery.
Most people looked for clues within the immediate vicinity of the gene but came up empty-handed. When Witcher extended his search further upstream, however, he discovered a binding site for CTCF, short for CCCTC-binding factor, which forms the centerpiece of the molecular fence posts that separate heterochromatin from the rest of the genome. “We found that the binding of this protein is lost from several binding sites in numerous types of cancer cells, leading to the collapse of the molecular boundary,” he says. “Once the boundary was gone, the adjacent heterochromatin encroached and silenced the nearest gene.”
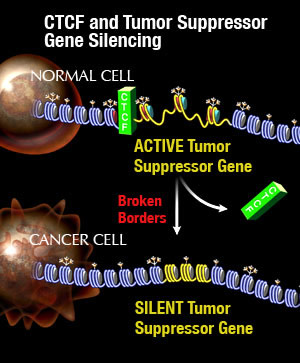
CTCF and Tumor Suppressor Gene Silencing
Image: Courtesy of Jamie Simon, Salk Institute for Biological Studies
Further experiments revealed that CTCF was missing because it lacked a chemical modification known as “PARlation,” lab lingo for poly(ADP-ribosyl)ation, which allows the protein to bind to select sites in the genome. “Without PARlation, CTCF fails to form the complex necessary to regulate p16 and the tumor suppressor RASSF1A and possibly others, explaining why breast cancer cells always contain both silenced p16 and silenced RASSF1A,” says Witcher.
“We believe that destabilization of specific chromosomal boundaries or loss of molecular fences through aberrant CTCF function may be a general mechanism to inactivate tumor suppressor genes and initiate tumorigenesis in numerous forms of human cancers,” says Emerson.
For information on the commercialization of this technology, please contact the Salk Office of Technology Development (858) 453-4100, Ext. 1278.
This work was supported by the Samuel Waxman Cancer Research Foundation and the Canadian Institute of Health Research.
About the Salk Institute for Biological Studies:
The Salk Institute for Biological Studies is one of the world’s preeminent basic research institutions, where internationally renowned faculty probe fundamental life science questions in a unique, collaborative, and creative environment. Focused on both discovery and mentoring future generations of researchers, Salk scientists make groundbreaking contributions to our understanding of cancer, aging, Alzheimer’s, diabetes, and cardiovascular disorders by studying neuroscience, genetics, cell and plant biology, and related disciplines.
Faculty achievements have been recognized with numerous honors, including Nobel Prizes and memberships in the National Academy of Sciences. Founded in 1960 by polio vaccine pioneer Jonas Salk, M.D., the Institute is an independent nonprofit organization and architectural landmark.
Office of Communications
Tel: (858) 453-4100
press@salk.edu