
July 2, 2015
Researchers have developed antibodies to help study critical chemical modifications responsible for a protein's development
Researchers have developed antibodies to help study critical chemical modifications responsible for a protein's development
LA JOLLA–Unveiling how the 20,000 or so proteins in the human body work–and malfunction–is the key to understanding much of health and disease. Now, Salk researchers developed a new technique that allows scientists to better understand an elusive step critical in protein formation.
The new method, described on July 2, 2015 in the journal Cell, allows researchers to map critical chemical tags–called phosphates–that bond to amino acids (the building blocks of proteins) in the final stages of creating a protein.
When a cell produces a protein, molecular synthetic machinery first pieces together the amino acids into a long strand that bends and folds as it lengthens. Enzymes then gather around the structure to make final tweaks–trimming the protein or adding chemical tags. Among these last modifications is phosphorylation–or addition of phosphate–of individual amino acids to change the protein’s function. Until now, pinpointing exactly where (and why) all those phosphates are added has been tricky.
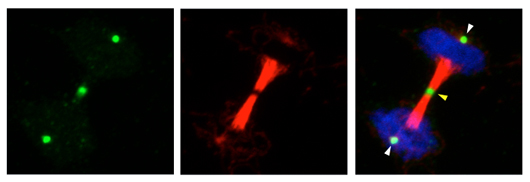
Salk scientists developed antibodies that tag essential chemicals involved in protein action. In this dividing cell, the new antibody (green) marks discrete areas of a critical but elusive chemical reaction that happens when cells divide (DNA in blue and tubulin, a protein needed for cell division, in red).
Click here for a high-resolution image.
Image: Courtesy of the Salk Institute for Biological Studies
“We know that 9 out of the 20 amino acids can be phosphorylated, but we know very little about most of them because they’re so hard to study,” says Tony Hunter, American Cancer Society Professor, holder of the Dulbecco Chair in the Salk’s Molecular and Cell Biology Laboratory and senior author of the new paper.
When phosphate is added to three particular amino acids–serine, threonine and tyrosine–it forms a strong chemical bond. Researchers can easily identify the location of these phosphates. But when the other six amino acids are phosphorylated, the phosphate is only loosely attached to each amino acid, causing problems for researchers trying to glean what happens in these cases. One of these amino acids, called histidine (or phosphohistidine once a phosphate is added), has been particularly tough to study.
“With those strong phosphorylation events, you can label cells, isolate proteins and analyze the proteins in various ways to find out where the phosphates are,” says Hunter. “You can’t do that with phosphohistidine because it’s so unstable it falls apart as you’re trying to isolate the proteins.”
Hunter and his collaborators realized that to study this unstable interaction, they would have to use a trick to make a stronger bond between phosphate and histidine. So they used a special kind of modified phosphate, called phosphonate, engineered to bind more tightly to either of the two spots on a histidine amino acid where phosphate can be added. Then, they developed antibodies that specifically recognize these stable phosphohistidine analogues, but also detect authentic phosphohistidine in proteins.
To test these new tools, the team added their phosphohistidine antibodies to a collection of different mammalian cells grown on slides and observed where in the cell the antibodies bound, which indicates parts of cells that have high levels of proteins with phosphohistidines.
“The thing that surprised us most is that when we stained the cells with the new antibodies, we saw discrete areas within the cells that had high levels of histidine phosphorylation, particularly when they were undergoing mitosis, the stage at which cells divide into two daughter cells,” says Hunter. They don’t yet know exactly why that is, but plan to continue to explore these results as well as detect the phosphorylation of other amino acids.
The team expects these antibody tools to be useful to other labs aiming to determine whether proteins of interest have any phosphohistidines.
The new method is “fairly easy for any lab to use,” Hunter says. “It doesn’t require a special instrument or anything, so I think it may be fairly quickly adopted.”
Other researchers on the study were Stephen Rush Fuhs, Jill Meisenhelder, Aaron Aslanian, Li Ma, Anna Zagorska and Greg Lemke, of the Salk Institute; Magda Stankova, Alan Binnie, Fahad Al-Obeidi and Jacques Mauger, of Sanofi; and John R. Yates III of The Scripps Research Institute.
The work and the researchers involved were supported by the National Institutes of Health, a Salk Institute Innovation Grant and the Helmsley Center for Genomic Medicine.
JOURNAL
Cell
AUTHORS
Stephen Rush Fuhs, Jill Meisenhelder, Aaron Aslanian, Li Ma, Anna Zagorska, Greg Lemke and Tony Hunter, of the Salk Institute; Magda Stankova, Alan Binue, Fahad Al-Obeidi and Jacques Mauger, of Sanofi; and John R. Yates III of the Scripps Research Institute
Office of Communications
Tel: (858) 453-4100
press@salk.edu
Unlocking the secrets of life itself is the driving force behind the Salk Institute. Our team of world-class, award-winning scientists pushes the boundaries of knowledge in areas such as neuroscience, cancer research, aging, immunobiology, plant biology, computational biology and more. Founded by Jonas Salk, developer of the first safe and effective polio vaccine, the Institute is an independent, nonprofit research organization and architectural landmark: small by choice, intimate by nature, and fearless in the face of any challenge.