
July 27, 2017
Researchers image 3D genome in nucleus of living human cell for the first time
Researchers image 3D genome in nucleus of living human cell for the first time
LA JOLLA—Stretched out, the DNA from all the cells in our body would reach Pluto. So how does each tiny cell pack a two-meter length of DNA into its nucleus, which is just one-thousandth of a millimeter across?
The answer to this daunting biological riddle is central to understanding how the three-dimensional organization of DNA in the nucleus influences our biology, from how our genome orchestrates our cellular activity to how genes are passed from parents to children.
Now, scientists at the Salk Institute and the University of California, San Diego, have for the first time provided an unprecedented view of the 3D structure of human chromatin—the combination of DNA and proteins—in the nucleus of living human cells.
In the tour de force study, described in Science on July 27, 2017, the Salk researchers identified a novel DNA dye that, when paired with advanced microscopy in a combined technology called ChromEMT, allows highly detailed visualization of chromatin structure in cells in the resting and mitotic (dividing) stages. By revealing nuclear chromatin structure in living cells, the work may help rewrite the textbook model of DNA organization and even change how we approach treatments for disease.
"One of the most intractable challenges in biology is to discover the higher-order structure of DNA in the nucleus and how is this linked to its functions in the genome," says Salk Associate Professor Clodagh O’Shea, a Howard Hughes Medical Institute Faculty Scholar and senior author of the paper. "It is of eminent importance, for this is the biologically relevant structure of DNA that determines both gene function and activity."
Click here for a high-resolution image.
Credit: Salk Institute
Ever since Francis Crick and James Watson determined the primary structure of DNA to be a double helix, scientists have wondered how DNA is further organized to allow its entire length to pack into the nucleus such that the cell's copying machinery can access it at different points in the cell's cycle of activity. X-rays and microscopy showed that the primary level of chromatin organization involves 147 bases of DNA spooling around proteins to form particles approximately 11 nanometers (nm) in diameter called nucleosomes. These nucleosome "beads on a string" are then thought to fold into discrete fibers of increasing diameter (30, 120, 320 nm etc.), until they form chromosomes. The problem is, no one has seen chromatin in these discrete intermediate sizes in cells that have not been broken apart and had their DNA harshly processed, so the textbook model of chromatin's hierarchical higher-order organization in intact cells has remained unverified.
To overcome the problem of visualizing chromatin in an intact nucleus, O’Shea’s team screened a number of candidate dyes, eventually finding one that could be precisely manipulated with light to undergo a complex series of chemical reactions that would essentially “paint” the surface of DNA with a metal so that its local structure and 3D polymer organization could be imaged in a living cell. The team partnered with UC San Diego professor and microscopy expert Mark Ellisman, one of the paper's coauthors, to exploit an advanced form of electron microscopy that tilts samples in an electron beam enabling their 3D structure to be reconstructed. By combining their chromatin dye with electron-microscope tomography, they created ChromEMT.
The team used ChromEMT to image and measure chromatin in resting human cells and during cell division when DNA is compacted into its most dense form—the 23 pairs of mitotic chromosomes that are the iconic image of the human genome. Surprisingly, they did not see any of the higher-order structures of the textbook model anywhere.

Click here for a high-resolution image
Credit: Salk Institute
"The textbook model is a cartoon illustration for a reason," says Horng Ou, a Salk research associate and the paper's first author. "Chromatin that has been extracted from the nucleus and subjected to processing in vitro—in test tubes—may not look like chromatin in an intact cell, so it is tremendously important to be able to see it in vivo."
What O'Shea's team saw, in both resting and dividing cells, was chromatin whose "beads on a string" did not form any higher-order structure like the theorized 30 or 120 or 320 nanometers. Instead, it formed a semi-flexible chain, which they painstakingly measured as varying continuously along its length between just 5 and 24 nanometers, bending and flexing to achieve different levels of compaction. This suggests that it is chromatin's packing density, and not some higher-order structure, that determines which areas of the genome are active and which are suppressed.
With their 3D microscopy reconstructions, the team was able to move through a 250 nm x 1000 nm x 1000 nm volume of chromatin's twists and turns, and envision how a large molecule like RNA polymerase, which transcribes (copies) DNA, might be directed by chromatin's variable packing density, like a video game aircraft flying through a series of canyons, to a particular spot in the genome. Besides potentially upending the textbook model of DNA organization, the team's results suggest that controlling access to chromatin could be a useful approach to preventing, diagnosing and treating diseases such as cancer.
"We show that chromatin does not need to form discrete higher-order structures to fit in the nucleus," adds O'Shea. "It's the packing density that could change and limit the accessibility of chromatin, providing a local and global structural basis through which different combinations of DNA sequences, nucleosome variations and modifications could be integrated in the nucleus to exquisitely fine-tune the functional activity and accessibility of our genomes."
Future work will examine whether chromatin's structure is universal among cell types or even among organisms.
Other authors included Sébastien Phan, Thomas Deerinck and Andrea Thor of the UC San Diego.
The work was largely funded by the W. M. Keck Foundation, the NIH 4D Nucleome Roadmap Initiative and the Howard Hughes Medical Institute, with additional support from the William Scandling Trust, the Price Family Foundation and the Leona M. and Harry B. Helmsley Charitable Trust.
JOURNAL
Science
AUTHORS
Horng D. Ou, Sébastien Phan, Thomas J. Deerinck, Andrea Thor, Mark H. Ellisman, Clodagh C. O'Shea
JOURNAL COVER

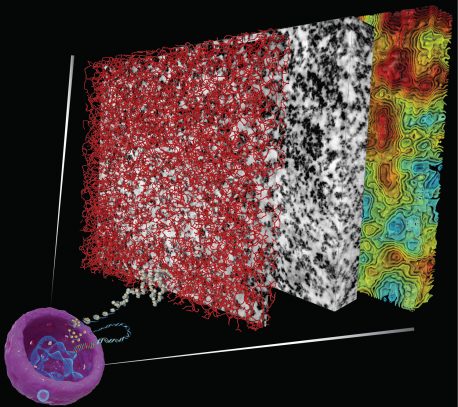
COVER Conceptual illustration of chromatin inside a cell's nucleus. To fit in the nucleus and mitotic chromosomes, DNA is assembled into chromatin, the structure of which determines the functions of our genome. By painting cells with a DNA binding dye that deposits a metal dusting on the surface of DNA, Ou et al. were able to visualize the chromatin structure of the human genome. See eaag0025. For more on the process behind the cover image, see http://scim.ag/2tUczje Illustration: Valerie Altounian/Science
Office of Communications
Tel: (858) 453-4100
press@salk.edu
Unlocking the secrets of life itself is the driving force behind the Salk Institute. Our team of world-class, award-winning scientists pushes the boundaries of knowledge in areas such as neuroscience, cancer research, aging, immunobiology, plant biology, computational biology and more. Founded by Jonas Salk, developer of the first safe and effective polio vaccine, the Institute is an independent, nonprofit research organization and architectural landmark: small by choice, intimate by nature, and fearless in the face of any challenge.