Associate Professor
Gene Expression Laboratory
Helen McLoraine Developmental Chair

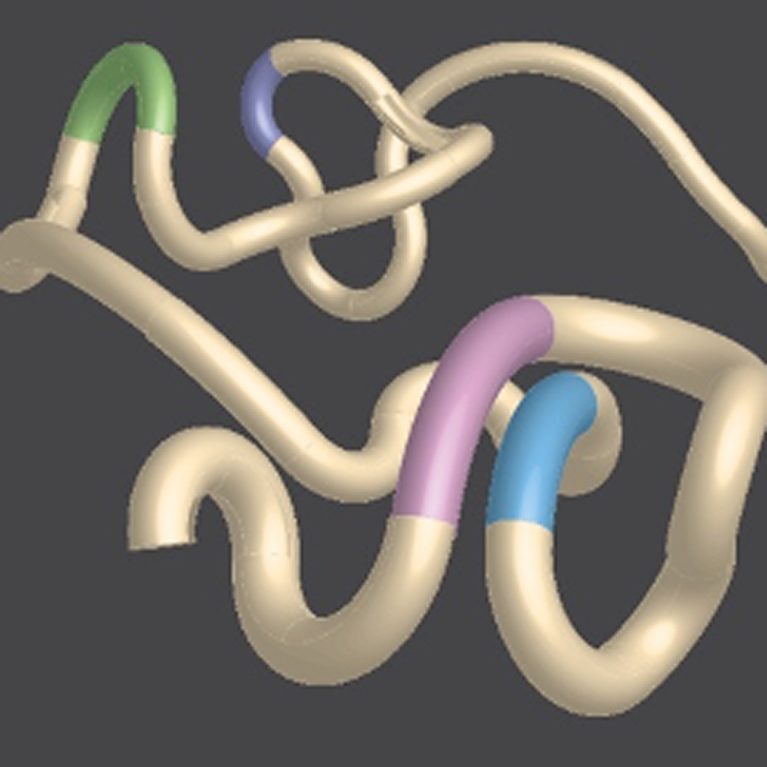
The human genome, the DNA blueprint for life, is organized in three-dimensional space inside of cells. While only two percent of the genome codes for proteins, much of the other 98 percent can serve a regulatory purpose, dictating when genes are expressed. The organization of these noncoding regions plays a critical role for proper gene regulation, yet understanding how genomes are folded and the consequences of folding errors are two extraordinary challenges for scientists.
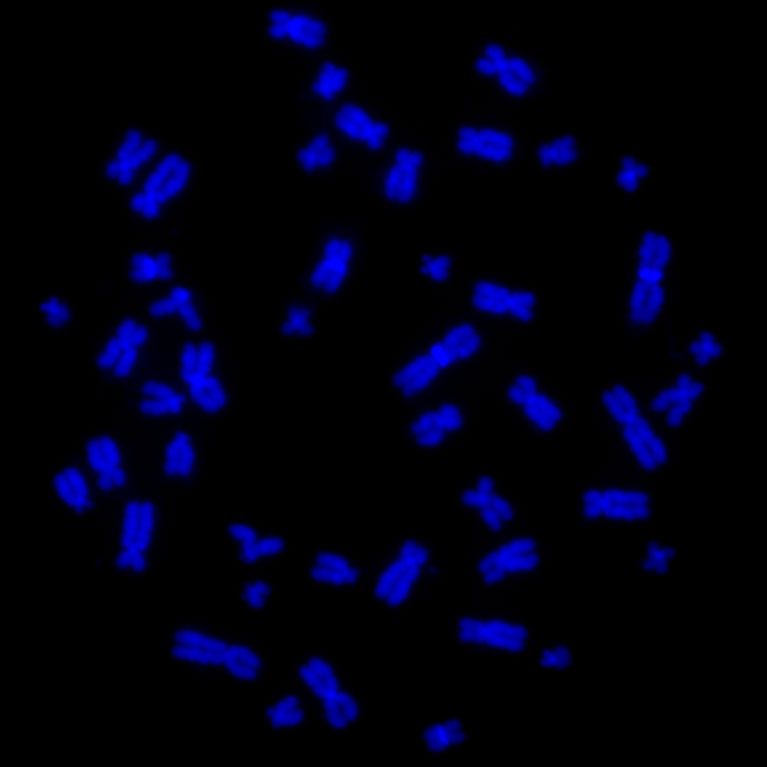
Dixon uses molecular and computational biology to explore how abnormal genome folding leads to errors in critical stretches of noncoding DNA that cause many diseases, such as cancer. His team is also developing new methods to study gene organization and gene function in single cells. By profiling each individual cell, the scientists gain extremely detailed (“high-resolution”) information about the different genes in each cellular system as well as insights into the molecular mutations that lead to disease.
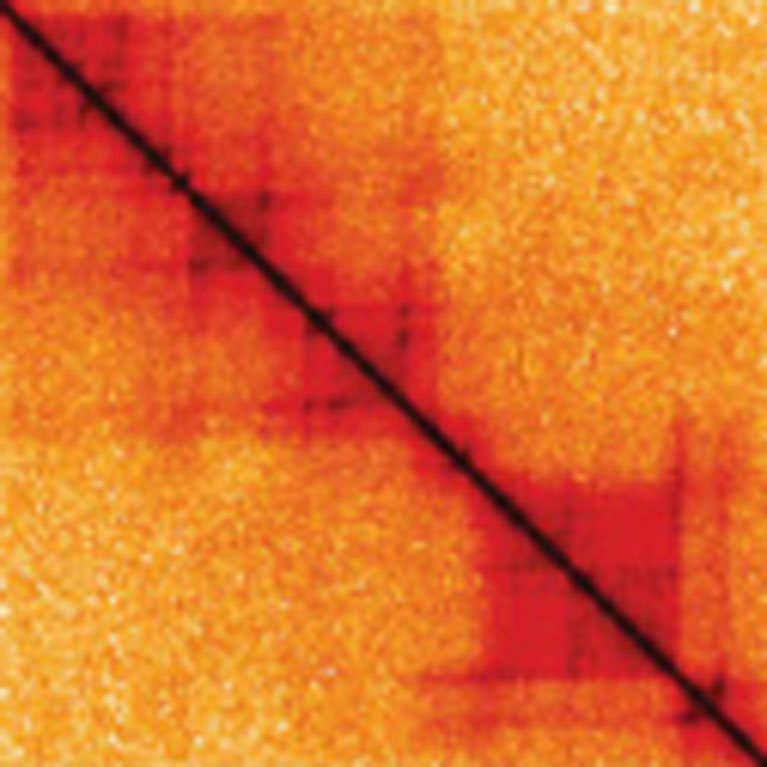
Dixon’s team described a fundamental feature of how genomes are organized, called topologically associating domains (TADs). These TADs act as genomic “neighborhoods” and function together in units.
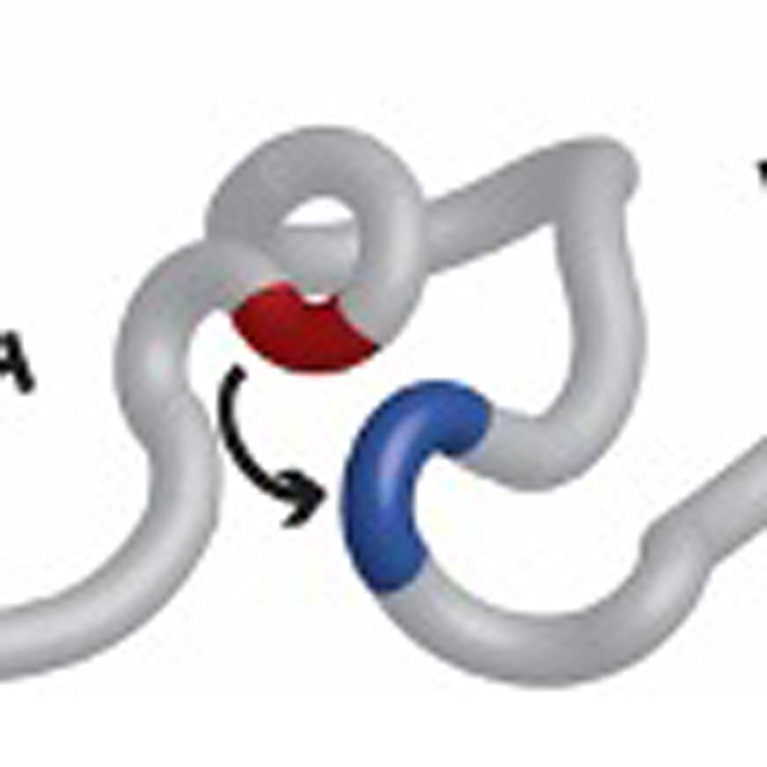
Gene mutations can break and rearrange chromosomes, which carry genetic information in the form of genes. Dixon discovered some of the basic consequences of these mutations on genome folding that lead to cancer.
Dixon created software and other genome technology tools that allow scientists to extract more genome sequence information than traditional techniques.
AB, Princeton University
PhD, University of California San Diego
MD, University of California San Diego
Helmsley-Salk Fellow, Salk Institute for Biological Studies